Aidan Baraban
A neuroscience major, Aidan will contribute to a collaborative project with the Henn Lab to characterize genes/variants implicated in skin pigmentation variation identified across individuals of South African ancestry using zebrafish.
Your Custom Text Here
Aidan Baraban
A neuroscience major, Aidan will contribute to a collaborative project with the Henn Lab to characterize genes/variants implicated in skin pigmentation variation identified across individuals of South African ancestry using zebrafish.
A historical scientific milestone has been achieved: the first complete sequence of a human genome (!) published in a special issue of Science. Scientists of the Telomere-to-Telomere (T2T) Consortium sequenced a fully homozygous human cell line, CHM13, generated by the erroneous fertilization of an enucleated egg, to reconstruct the complete genome. The new assembly, named T2T-CHM13, includes 8% of heterochromatin completely missing from the previous human genome assembly (GRCh38). Besides the original Human Genome Project, this is the second most significant progress in our sequencing and understanding of the human genome.

Summary abstract from “A complete reference genome improves analysis of human genetic variation”
Dennis lab members, including Megan and graduate students Colin Shew and Daniela Soto, contributed to a number of the published studies, including:
“A complete reference genome improves analysis of human genetic variation,” where the impact of the complete genome in the analysis of human genetic variation was thoroughly assessed. This was a truly collaborative effort led by four labs (Schatz, McCoy, Zook, and ours). In particular, Daniela performed genome-wide analysis of collapsed duplication in GRCh38, finding 8 Mbp of sequence incorrectly represented in GRCh38 and fixed in T2T-CHM13. Further, we helped demonstrate significant improvements in variant calling across medically-relevant genes.
“Complete genomic and epigenetic maps of human centromeres” by generating CHM13 RNA-seq data, with Colin analyzing transcript abundance across the T2T-CHM13 transcriptome and showing expression of numerous genes near centromeres.
“The complete sequence of a human genome”, with Megan performing comparative genomic analysis of the expanded FRG1 primate-specific gene expansion, which has several missing copies in GRCh38 that are resolved in T2T-CHM13.
This new genome opens many possibilities for scientists who can now explore the most repetitive sequence of the human genome, such as centromeres. Particularly interesting for us is the full resolution of segmental duplications, large historical duplicated segments that are a hallmark of great ape evolution and harbor genes associated with neurodevelopment. Moving forward, we are fully embracing T2T-CHM13 to answer questions regarding the evolution and disease implication of human-specific duplicated genes at unprecedented resolution (and hope many of you will too!).
Additional resources available for the new T2T-CHM13 genome include:
CHM13 open-access data available via GitHub
Dennis lab analysis available via GitHub
Genome stratifications of reference artefacts (incl. collapsed dups) are here
You can read Megan’s commentary about the implications of this scientific achievement in a perspective published in Genome Research. Also, our work, in addition to contributions by Prof Chuck Langley, were highlighted by UC Davis here.
This past year, we have welcomed two new undergrads, Jeffrey Zang (Computer Science) and Gabriana La (Biochemistry), and a new grad student, Sean McGinty (Integrative Genetics & Genomics).
Gabriana was recently awarded the UC Davis Genome Center Charles and Nanci Cooper Undergraduate Award. Sean received a UC Davis Dean’s Distinguished Graduate Fellowship.
Welcome!!

Our recent paper “Evaluation of CRISPR gene-editing tools in zebrafish” led by grad student José Uribe-Salazar is out in BMC Genomics. Several others from the lab contributed including lab manager Gulhan Kaya, volunteer Aadithya Sekar, and postbacs/undergrads KaeChandra Weyenberg and Cole Ingamells.
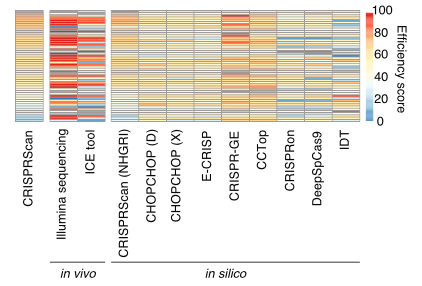
Some highlights:
We tested editing accuracy and predictability of 50 guide RNAs (from IDT) with SpCas9 using in vivo, in silico, and in vitro methods.
Large discrepancies existed between in silico editing predictive tools and empirical in vivo cutting efficiencies. The most accurate prediction resulted from using CRISPRScan against a reference specific to our zebrafish strain.
We observed low frequency of off-target mutations with no method (in silico or in vitro) effectively predicting these. More analyses are needed to understand the variability involved in off-target cutting events.
To characterize commonly-used controls in zebrafish CRISPR experiments, we evaluated larvae solely injected with SpCas9 (enzyme or mRNA) and found no evidence of an excess of somatic mutations in these injected larvae.
Performing RNA-seq of injection controls revealed notable variability in gene expression profiles compared to uninjected controls, including genes involved in several metabolic pathways that highlight a potential lasting effect of the microinjection. We recommend caution if performing characterizations of these particular genes in mosaic crispants.
Overall, our hope is for this resource to be helpful for the zebrafish/CRISPR community while they plan their future awesome experiments!

Our recent paper “RapID Cell Counter: Semi-automated and mid-throughput estimation of cell density within diverse cortical layers” was published in eNeuro co-led by IGG grad students Aarthi Sekar and Thiago Sanches and in collaboration with the amazing Simó lab at UC Davis. After struggling to manually count neurons from images of in utero electroporation of mouse neocortex, we decided to make an easy-to-use automated tool that can quantify fluorescently-labeled cells across defined layers. The method is called RapID and is available here.

Some highlights of our study:
RapID runs as an easy-to-use GUI, with step-by-step instructions on how to install and run as supplementary materials and on the Github. Our hope is that the tool will be useful to the community with install and implementation representing only a small hurdle. Please reach out to us if you encounter difficulties!
Users define parameters for images based on expected cell size, density, and brightness. RapID subsequently will quantify labeled cells within a user-defined quadrangle (split by set number of layers). Parameters used for diverse cell types by our groups are included in the study as a starting point for users.
We successfully show that RapID results match manual “click” identification using Fiji’s Cell Counter and apply it to diverse cell images, including astrocytes and dopaminergic neurons.
As an added bonus: the beautiful images of dopaminergic neurons, made by the talented Keiko Hino and Sergi Simó, were selected as an official Zoom backdrop for the Society for Neuroscience available for download on their website!

We are so proud of former Dennis lab undergrads heading off to grad school this fall!

Matangi Kumar will be starting in the Vision Science Ph.D. program at UC Berkeley. She also received a UC Davis Distinguished Scholar award as one of the top Genetics and Genomics undergrads in her graduating class!
Mira Mastoras is joining the Biomolecular Engineering and Bioinformatics Ph.D. program at UC Santa Cruz.
Eva Ferino was accepted to the Genetic Counseling master’s program at UCSF.

Welcome to the newest member of the lab Tasha Mariano joining via the PREP@UCD program! Tasha will use zebrafish to characterize human-specific duplicated genes and their role in brain evolution.
Our recent study “Diverse Molecular Mechanisms Contribute to Differential Expression of Human Duplicated Genes” was published in Molecular Biology and Evolution led by IGG grad student Colin Shew, with support from various current/former Dennis lab members Paulina Carmona-Mora, Daniela Soto, Mira Mastoras, Elizabeth Roberts, Joseph Rosas, and Gulhan Kaya and collaborator Henriette O’Geen. Using diverse functional genomic datasets, we characterized expression of recent human gene duplicates to better understand their functional fates and mechanisms underlying altered paralog expression.

Major highlights of the study:
Most human derived duplicate genes diverge in expression from their human and chimp counterparts, suggesting they’re undergoing neofunctionalization or pseudogenization.
In some cases derived genes may have usurped function of their ancestral paralogs or function through increased gene dosage.
Evolutionary age, post-transcriptional mechanisms, copy-number, and truncation status cannot completely explain observed diverged expression of genes in lymphoblastoid cell lines
ChIP-seq datasets miss most histone marks in duplicated regions that we recover using a modified bioinformatic pipeline that allows multiple read mappings.
We did not detect global differences in regulatory signatures between ancestral and derived paralogs, but rather, individual differences in cis regulatory elements, which we validate using reporter assays.

Hooray for Colin for his recent NRSA from NHGRI to characterize gene regulation of duplicated genes!
Tha lab’s first zebrafish paper “Assessment of Autism Zebrafish Mutant Models Using a High-Throughput Larval Phenotyping Platform” was published in a special topic of Frontiers in Cell and Developmental Biology: Zebrafish Models for Human Disease Studies led by postdoc Alexandra Colón-Rodríguez with support from present and former Dennis lab members José Uribe Salazar, KaeChandra Weyenberg, Aditya Sriram, Alejandra Quezada, Gulhan Kaya, Emily Jao, and Brittany Radke, as well as collaborator Pam Lein. Amazingly, co-authors included three undergrads and a high school student!
Some highlights:
Tested impacts on larval development of a high-throughput platform allowing morphological measurements using the VAST imaging system coupled with motion-tracking using DanioVision to detect drug-induced seizures.
Identified developmental defects (altered brain and eye sizes) and enhanced seizures for CRISPR mutants targeting orthologs of autism genes, SYNGAP1 and SLC7A5.
Established defects could be detected in both mosaic and stable mutant lines, opening up the possibility to increase throughput of genes screened.


The Dennis lab is partying over news that IGG grad student José was awarded this prestigious ASHG award for his research on our human-duplicated gene SRGAP2 zebrafish model. Congrats José!

We are excited to welcome new Autism Research Training Program postdoc Sierra Nishizaki to characterize genes implicated in autism and megalencephaly using human sequence datasets and zebrafish

Graduate student Daniela Soto recently gave a webinar via Oxford Nanopore Technologies on our recent publication of chimpanzee structural variants (SVs; a joint co-authored project with graduate student Colin Shew). If you are interested in learning how to apply ONT and optical mapping reads to discover novel SVs and didn’t get a chance to check it out live, the recording is available here.

The Dennis lab wants to congratulate junior specialist KaeChandra Weyenberg as she departs to pursue her master’s degree in Public Health from East Tennessee University.
We are also happy to send off graduating undergrads Aditya Sriram and Elizabeth Roberts. Aditya is headed to the University of Washington to pursue a master’s degree in Epidemiology and Elizabeth will pursue research at the Broad Institute in the Lander lab as she prepares to apply to grad school in the future.
We are also celebrating the news of several former Dennis lab undergrads:
Maram Bader has accepted an offer of admission to San Jose State University’s master’s in Microbiology and Molecular Biology program.
Dhriti Jagannathan plans to join the Genetic Counseling master’s program at the University of Minnesota.
Juliann Wang will start medical school at the University of Alabama at Birmingham School of Medicine.